Partnering for progress: A decade of trust in Devyser CFTR
With the IVDR transition approaching, European clinical laboratories are preparing for changes that...
What is Devyser CFTR?
Devyser CFTR is a CE-marked (IVDR) NGS solution for full-gene CFTR analysis that detects SNVs and indels, CNVs, and the poly‑T/TG repeat in one run. The streamlined protocol provides unmatched simplicity, minimizing handling and supporting standardized routine diagnostics. The assay has been designed to keep pace with evolving clinical guideline recommendations, for example the updated ACMG variant list and the ever-expanding CFTR2 database.1,2

.png?width=300&height=300&name=Milan-M.-photo_circle%20(1).png)
“We have tried many assays of various types, but targeted, amplicon-based sequencing with high quality and high coverage is the best method for a clearly defined disease like cystic fibrosis.”
Prof. Milan Macek (M.D., DSc.), President of the Czech Society of Medical Genetics and Genomics
Devyser CFTR – Product features
Equip your lab with IVDR-certified CFTR gene sequencing in a single tube: efficient, reliable, and accurate.
Pre-dispensed indexes and a single pooled clean‑up minimize handling and lower mix‑up risk; typical hands-on time is under 45 minutes.
Broad coverage
Targets include exons, exon–intron junctions, promoter, and selected deep‑intronic regions, reducing the need for downstream assays and supporting cost‑effective testing.
Amplicon Suite CFTR converts FASTQ into consolidated results for SNVs/indels, CNVs, and poly‑T/TG, minimizing reliance on in‑house bioinformatics. The software supports filtered results for both screening and full gene analysis.
Fixed mutation panels can show ancestry‑related bias, lowering sensitivity in many non‑European groups. Full‑gene sequencing mitigates missed or delayed diagnoses in underrepresented populations.
Population genetics and equity
Sequencing beyond legacy mutation panels aligns testing with today’s changing demographics and supports equitable carrier screening and diagnostics.
Clinical utility over panel bias
Broader CFTR genotyping captures both common and rare or population‑specific variants, improving clinical decision‑making.
Why choose Devyser CFTR?

Scalable solution
Suitable for both small laboratories and high‑throughput centers. From 8 to 192 samples per experiment, run urgent small batches to keep turn around time (TAT) short or fill plates for maximum efficiency with predictable cost per sample.


Simplest workflow
Single-tube target amplification to limit mix-ups and contamination, pre-dispensed indexing to reduce handling and errors, and pooled library clean-up in one tube to save time.
Contact us to learn more about Devyser CFTR.
Instructions for use
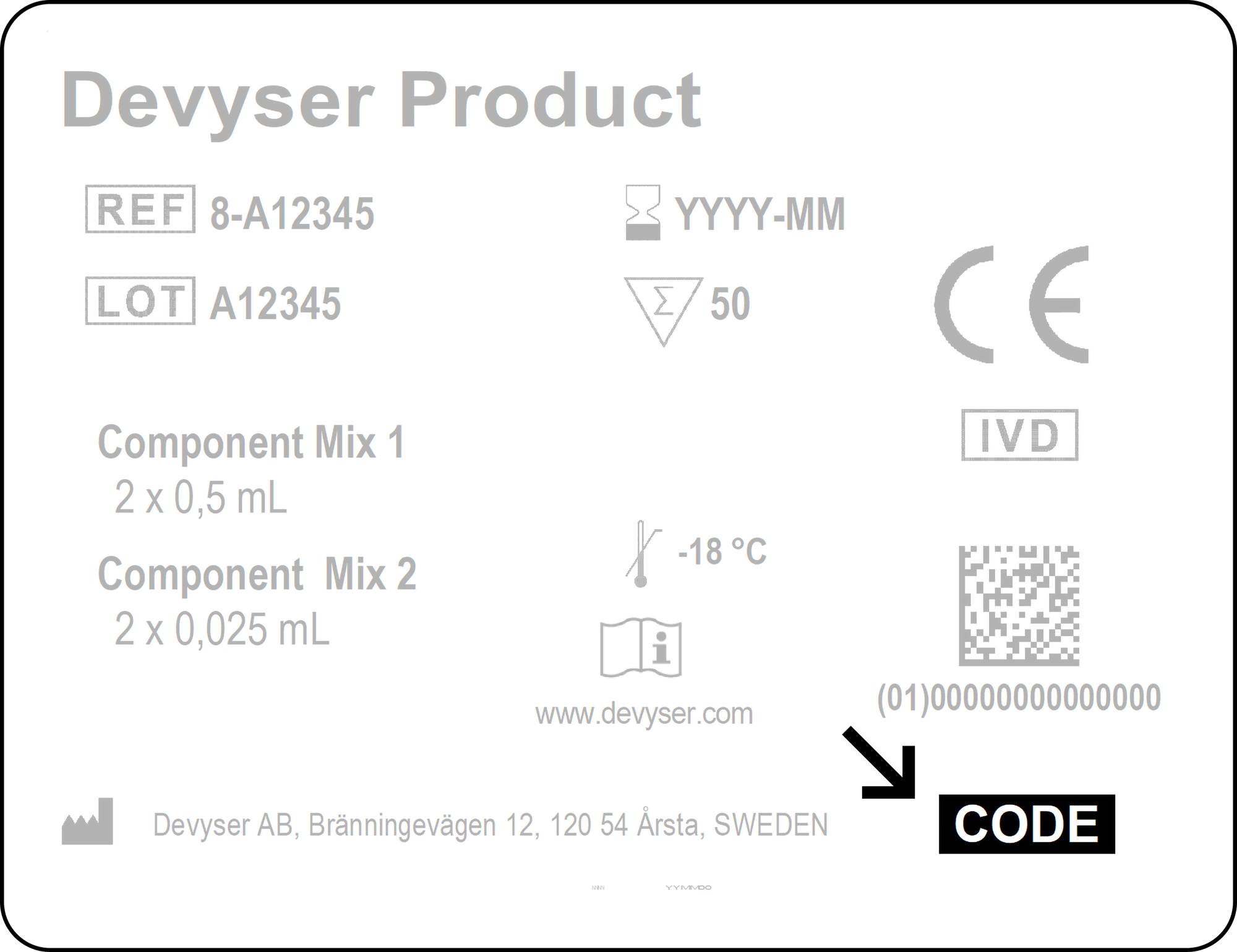
Enter access code found in the lower right corner of the label on the kit box.
Certificates
Download a specific Batch Release Certificate (BRC) below.
Guidelines and handbooks (1)
Product information (2)
Software settings (13)
Safety data sheets (1)
Data tables (0)
Enter access code found in the lower right corner of the label on the kit box.

Download a specific Batch Release Certificate (BRC) below.
CE-IVDR
Devyser CFTR NGS
Single‑tube full CFTR gene sequencing with comprehensive variant detection.
8-R101-24
8-R101-96
Pack size: 24
Pack size: 96
CE-IVD
Devyser CFTR NGS
Single‑tube full CFTR gene sequencing with comprehensive variant detection.
8-A101-8
8-A101-24
8-A101-96
Pack size: 8
Pack size: 24
Pack size: 96
RUO
Devyser CFTR NGS
Single‑tube full CFTR gene sequencing with comprehensive variant detection.
8-A103-8
8-A103-24
8-A103-96
Pack size: 8
Pack size: 24
Pack size: 96
How is Devyser CFTR different from other CFTR assays?
It is a single‑tube, amplicon‑based NGS assay with pre‑dispensed indexing and one pooled clean‑up to minimize handling, mix‑ups, and contamination. The CE‑marked (IVDR) workflow includes integrated analysis (Amplicon Suite CFTR) that reports SNVs, indels, CNVs, and poly‑T/TG from one run. A unique dual detection approach allows for robust detection of exon-spanning deletions.
How are results analyzed, and which variants are reported?
Amplicon Suite CFTR converts FASTQ files into consolidated calls for SNVs/indels, CNVs, and poly‑T/TG, reducing reliance on in‑house bioinformatics.
What sample types are accepted?
Extracted genomic DNA (refer to the IFU for full details and requirements).
How does Devyser CFTR integrate with our workflow?
Compatible with Illumina platforms; typical hands‑on time is under 45 minutes. Scales from urgent small batches to high throughput with 8 to 192 samples per index plate and predictable cost per sample.
Is the assay suitable for carrier screening programs?
Yes. Sequencing mitigates ancestry‑related bias inherent to fixed mutation panels and supports equitable testing in reproductive carrier screening.
Does Devyser CFTR include guideline‑recommended variants?
The assay is designed to align with evolving recommendations (e.g., ACMG) and developed with the CFTR2 database in mind. It detects common, rare, or population‑specific variants. For site‑specific requirements, consult the IFU and release notes.
Does Devyser offer technical and scientific support?
Yes. Support is available for assay setup and validation as well as data analysis and troubleshooting.
Devyser CFTR in literature
These peer‑reviewed studies and conference posters demonstrate how Devyser CFTR is applied in real‑world diagnostics and research-spanning variant detection, newborn screening, and population genetics across multiple countries.

With the IVDR transition approaching, European clinical laboratories are preparing for changes that...
Read More
-1.png)
We spoke with Dr. Andrew Purvis, Clinical Scientist for West of Scotland Centre for Genomic...
Read More
-2.png)
Devyser has secured a CFTR NGS proposal with UNC Hospitals. The proposal with UNC Hospitals is...
Read More
-1.png)
On October 26th, we hosted an insightful webinar by Prof. Milan Macek (M.D., DSc.), where he...
Read More